GSoC
GSoC.Rmd
#devtools::install_github("ZeroDawn0D/tRiad")
library(triad)Introduction
This package has been developed as part of Google Summer of Code 2022, with the help of my mentors Di Cook and Harriet Mason. It calculates the Standard Delaunay Triangulations of a set of 2D points using the Sloan(1987) algorithm.
Link to GitHub repository: ZeroDawn0D/tRiad
There are two ways to use triad:
C++ functions (faster but harder to read and modify)
The DelTri() function creates an object of class
triad. This object can be plotted.
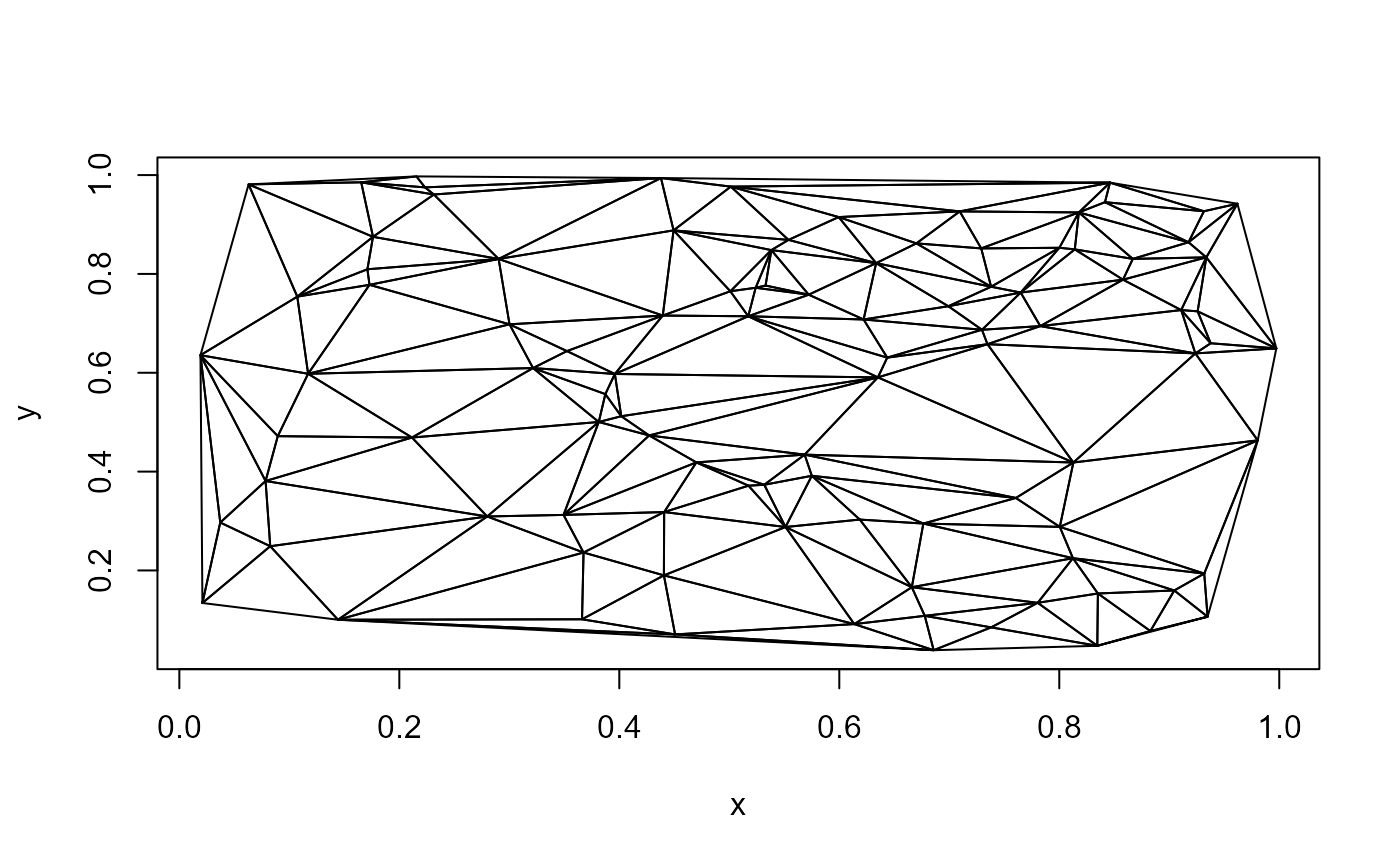
R functions (slower but easier to read and modify)
The del.tri() function does what DelTri()
does. The object can be plotted in a similar way.
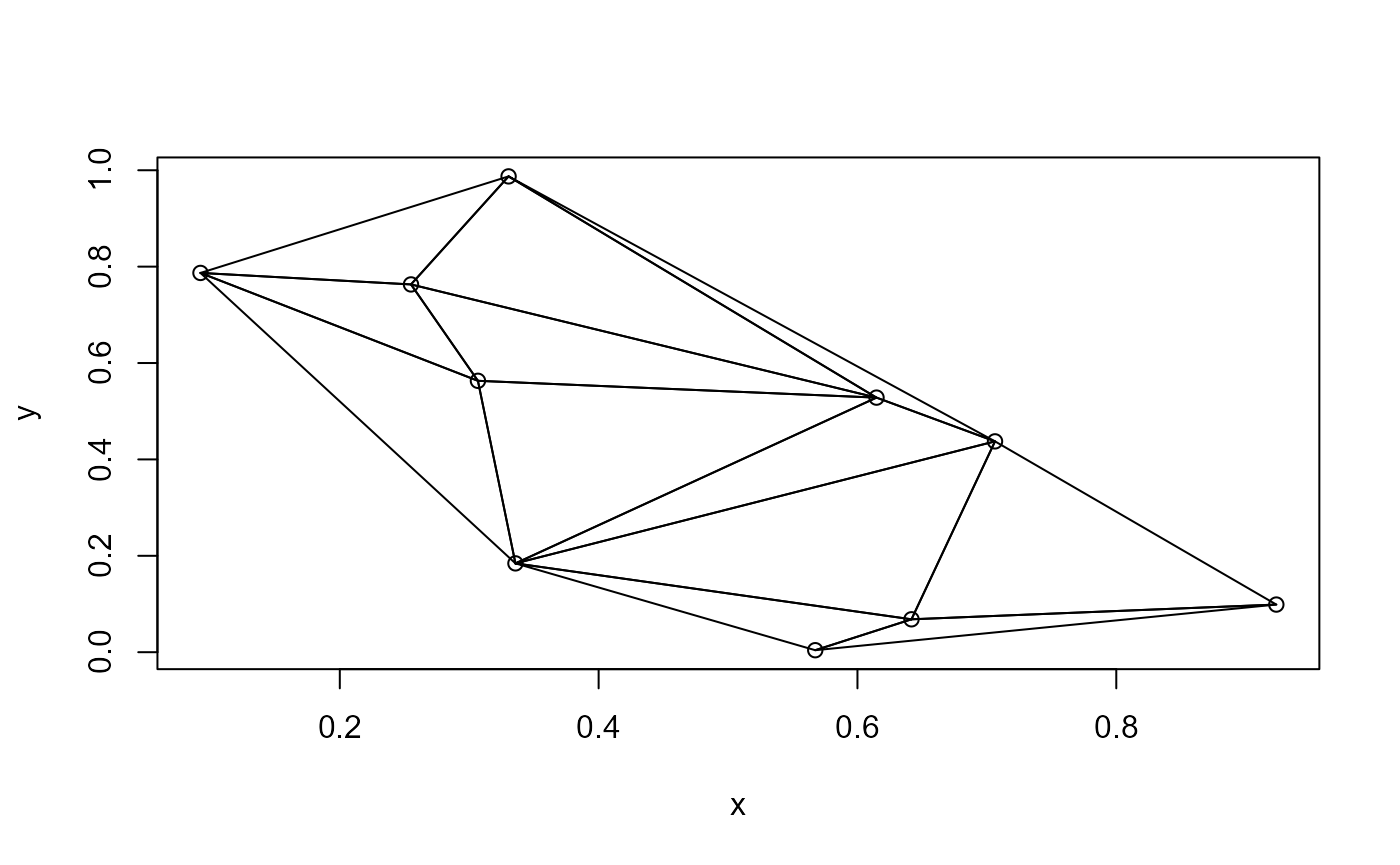
Time comparisons
We also compare with interp::tri.mesh()
#install.packages("interp")
library(interp)
#> Warning: package 'interp' was built under R version 4.2.1
x <- runif(500)
y <- runif(500)
system.time(triad::del.tri(x,y))
#> user system elapsed
#> 0.25 0.00 0.25
system.time(triad::DelTri(x,y))
#> user system elapsed
#> 0.02 0.00 0.02
system.time(interp::tri.mesh(x,y))
#> user system elapsed
#> 0.01 0.00 0.02For larger input:
system.time(triad::del.tri(x,y))
#> user system elapsed
#> 6.03 0.89 7.06
system.time(triad::DelTri(x,y))
#> user system elapsed
#> 0.31 0.00 0.31
system.time(interp::tri.mesh(x,y))
#> user system elapsed
#> 0.13 0.00 0.12SWAP subroutine
The code makes certain changes to the SWAP subroutine. For some reason, the algorithm as described in the paper broke when dealing with concave quadrilaterals.
swapR <- function(x1, y1, x2, y2, x3, y3, xp, yp){
x1p <- x1 - xp
y1p <- y1 - yp
x2p <- x2 - xp
y2p <- y2 - yp
dot.1p.2p <- x1p*x2p + y1p*y2p
mod1p <- sqrt(x1p*x1p + y1p*y1p)
mod2p <- sqrt(x2p*x2p + y2p*y2p)
cos.alpha <- dot.1p.2p / (mod1p * mod2p)
if(cos.alpha > 1){
cos.alpha <- 1
}
if(cos.alpha < -1){
cos.alpha <- -1
}
alpha <- acos(cos.alpha)
#angle between vector 3->1 and 3->2
x13 <- x1 - x3
y13 <- y1 - y3
x23 <- x2 - x3
y23 <- y2 - y3
dot.13.23 <- x13*x23 + y13*y23
mod13 <- sqrt(x13*x13 + y13*y13)
mod23 <- sqrt(x23*x23 + y23*y23)
cos.gamma <- dot.13.23/(mod13*mod23)
if(cos.gamma > 1){
cos.gamma <- 1
}
if(cos.gamma < -1){
cos.gamma <- -1
}
gamma <- acos(cos.gamma)
return((alpha+gamma) > pi)
}Further work
- Implementing a C++ version of the Constrained Delaunay
Triangulations.
tripackhas a FORTRAN implementation which is not open source -
plot.triad()is slower thanplot.triSh()although both are written in R. Optimise it even further - Optimise
DelTri()to be as fast asinterp::tri.mesh(), if not faster
Optimising C++ code even further
The code makes use of <vector.h> and
<stack.h> from the C++ Standard Template Library.
Currently, I use vector<vector<double>> to
represent 2D matrices, and ample use of push_back(). This
leads to a lot of resizing that could slow down the code. I tried to
reserve memory beforehand using reserve() but it became
even slower. I also tried pre-allocating the memory but it did not work.
There could be some way to use double** to optimise it.
This is currently a work in progress.